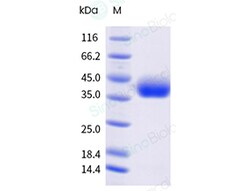
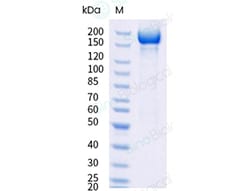
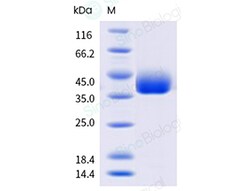
A DNA sequence encoding the SARS-CoV-2 (KP.2) Spike S1+S2 trimer (YP_009724390.1, with mutations ins16MPLF, T19I, R21T, L24del, P25del, P26del, A27S, S50L, H69del, V70del, V127F, G142D, Y144del, F157S, R158G, N211del, L212I, V213G, L216F, H245N, A264D, I332V, G339H, R346T, K356T, S371F, S373P, S375F, T376A, R403K, D405N, R408S, K417N, N440K, V445H, G446S, N450D, L452W, L455S, F456L, N460K, S477N, T478K, N481K, V483del, E484K, F486P, Q498R, N501Y, Y505H, E554K, A570V, D614G, P621S, H655Y, N679K, P681R, R682G, R683S, R685S, N764K, D796Y, F817P, A892P, A899P, S939F, A942P, Q954H, N969K, K986P, V987P, V1104L, P1143L and furin cleavage site mutants) was expressed with bacteriophage T4 fibritin and a C-terminal polyhistidine tag followed by an AVI tag. The mutations were identified in the SARS-CoV-2 variant (known as variant KP.2).The expressed protein was biotinylated in vivo by the Biotin-Protein ligase (BirA enzyme) which is co-expressed.